2025 Fall CTSA Program Annual Meeting

2025 Fall CTSA Program
Annual Meeting
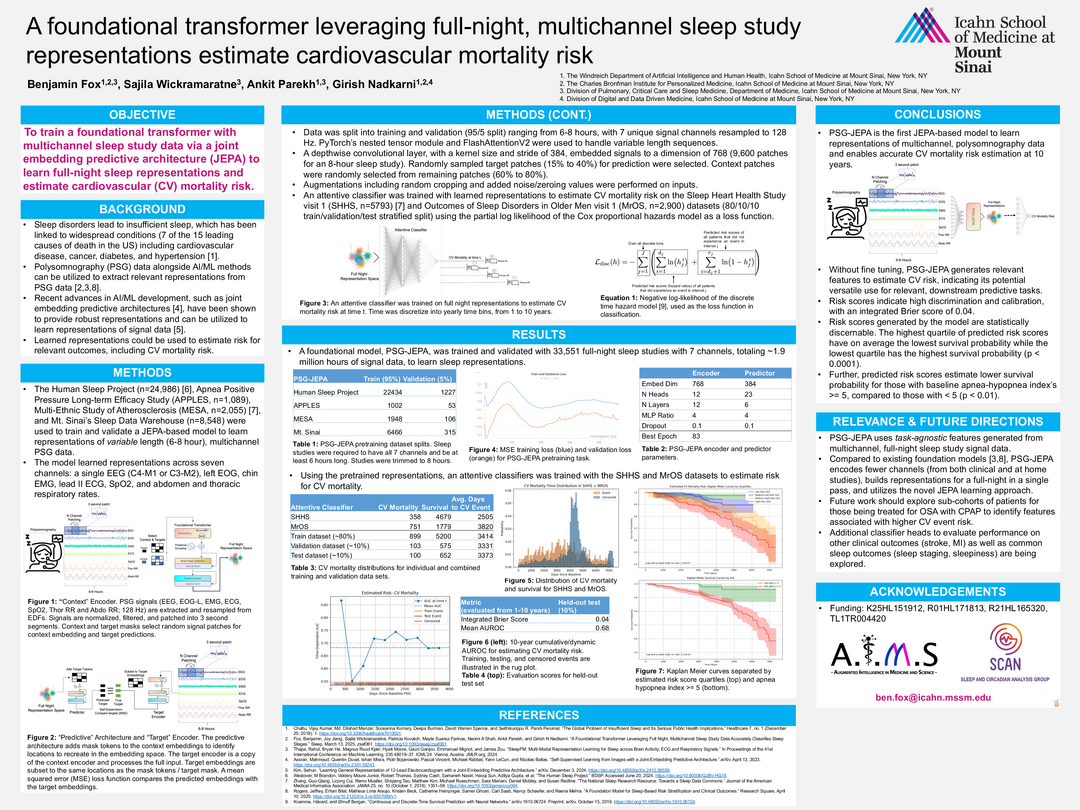
A foundational transformer leveraging full-night, multichannel sleep study representations estimate cardiovascular mortality risk
Benjamin Fox
Hub not available
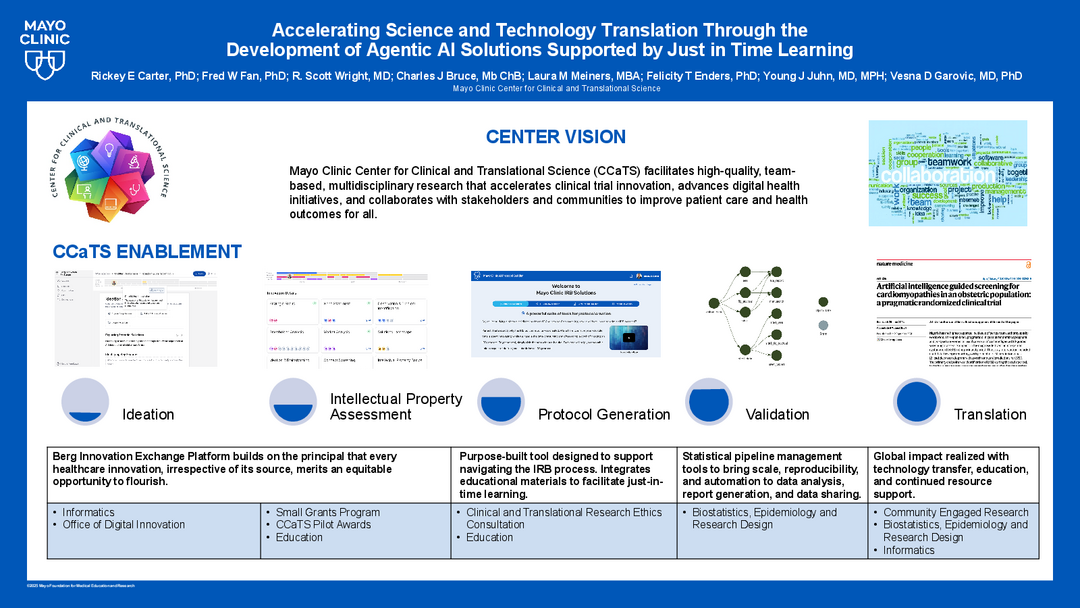
Accelerating Science and Technology Translation Through the Development of Agentic AI Solutions Supported by Just in Time Learning
Rickey Carter
Hub not available

Adapting and Disseminating the Blue Star Investigator Training Program: Variations on a Theme
Kris Markman
Hub not available
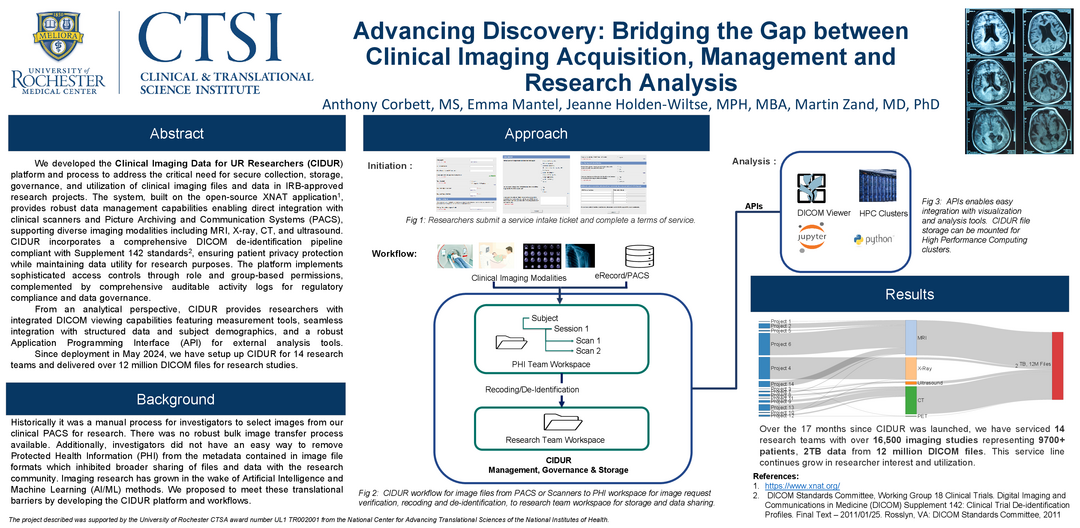
Advancing Discovery: Bridging the Gap between Clinical Imaging Acquisition, Management, and Research Analysis
Jeanne Holden-Wiltse
Hub not available

Advancing the Translational Science Workforce: An Integrated Model of Research, Education, Mentorship, and Community Engagement
Maija Williams
Hub not available
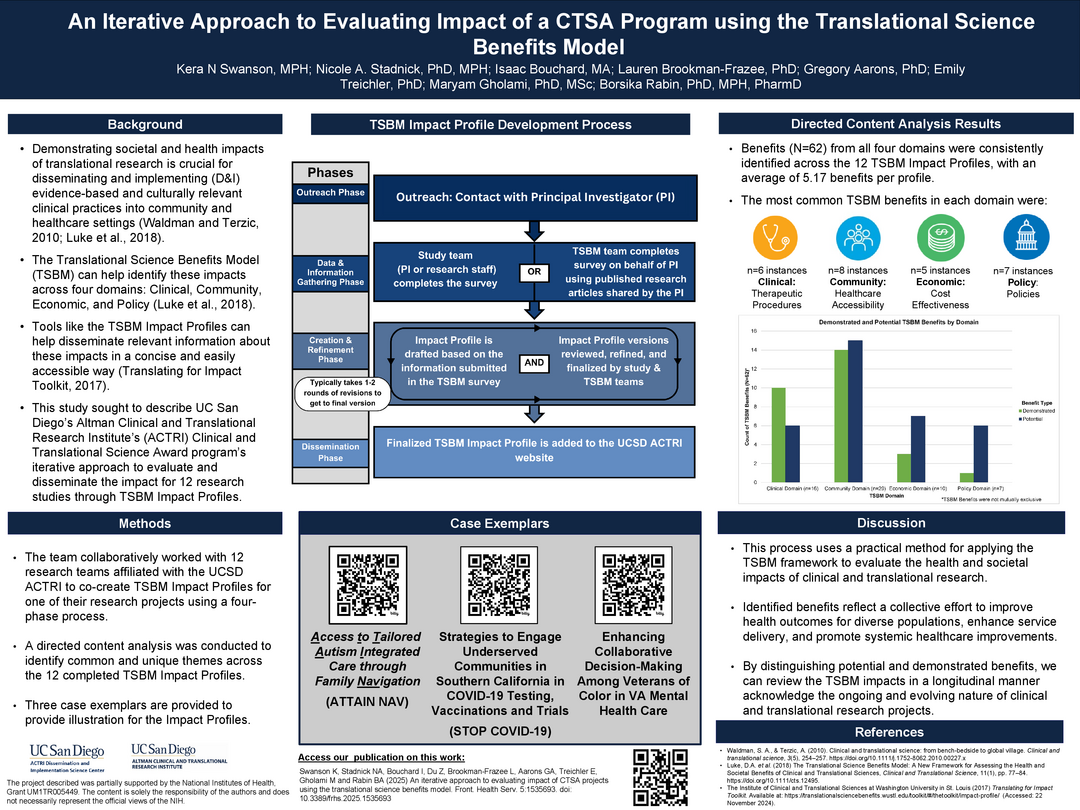
An iterative approach to evaluating impact of a CTSA program using the Translational Science Benefits Model
Borsika Rabin
Hub not available
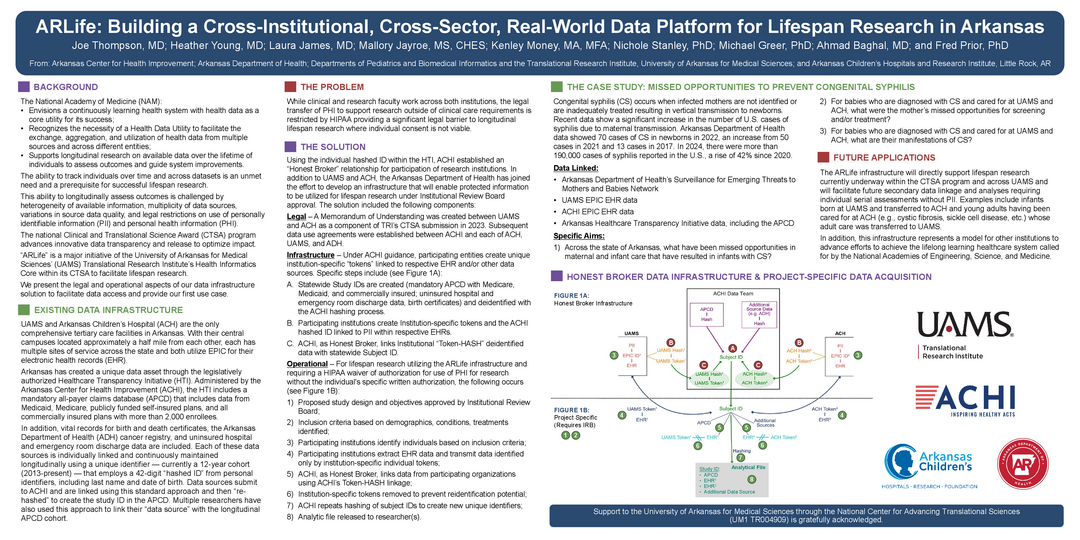
ARLife: Building a Cross-Institutional, Cross-Sector, Real-World Data Platform for Lifespan Research in Arkansas
Joe Thompson
Hub not available
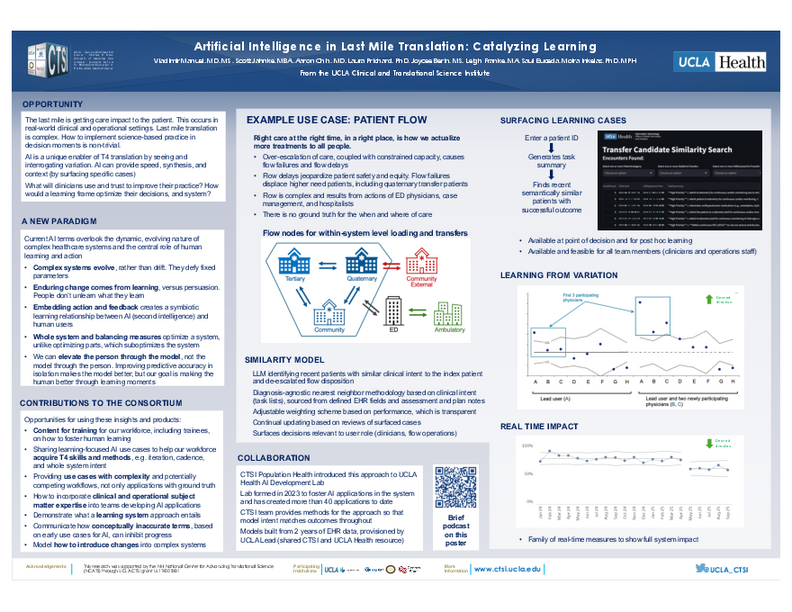
Artificial Intelligence in Last Mile Translation: Catalyzing Learning
Vladimir Manual
Hub not available
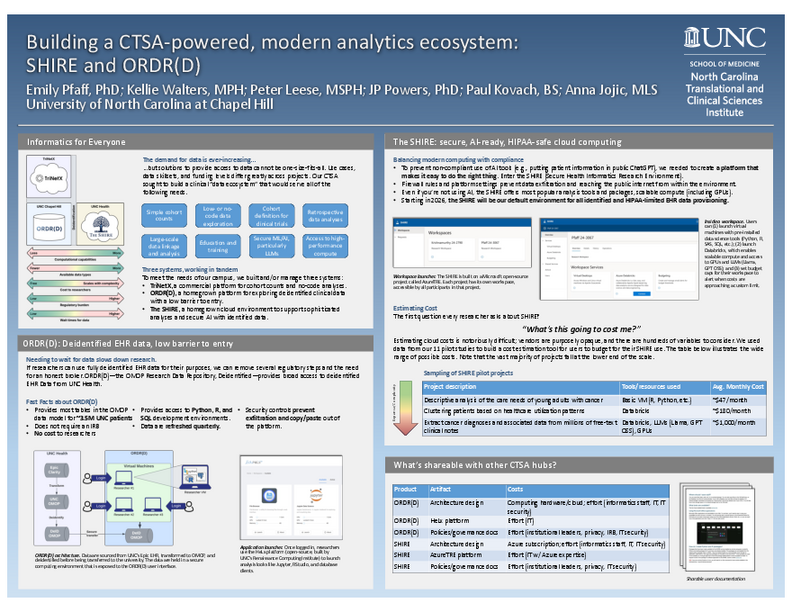
Building a CTSA-powered, modern analytic ecosystem: SHIRE and ORDR(D)
Emily Pfaff
Hub not available

Building a Digital Twin Model for Predicting Maternal Health Risks: Utilizing Multi-Institutional Collaborations to Advance Innovative Clinical and Translational Science
Alexander Libin
Hub not available



