Log in to view meeting materials such as meeting slides and recordings
2025 Fall CTSA Program Annual Meeting

2025 Fall CTSA Program
Annual Meeting
A foundational transformer leveraging full-night, multichannel sleep study representations estimate cardiovascular mortality risk
Hub not available
Abstract
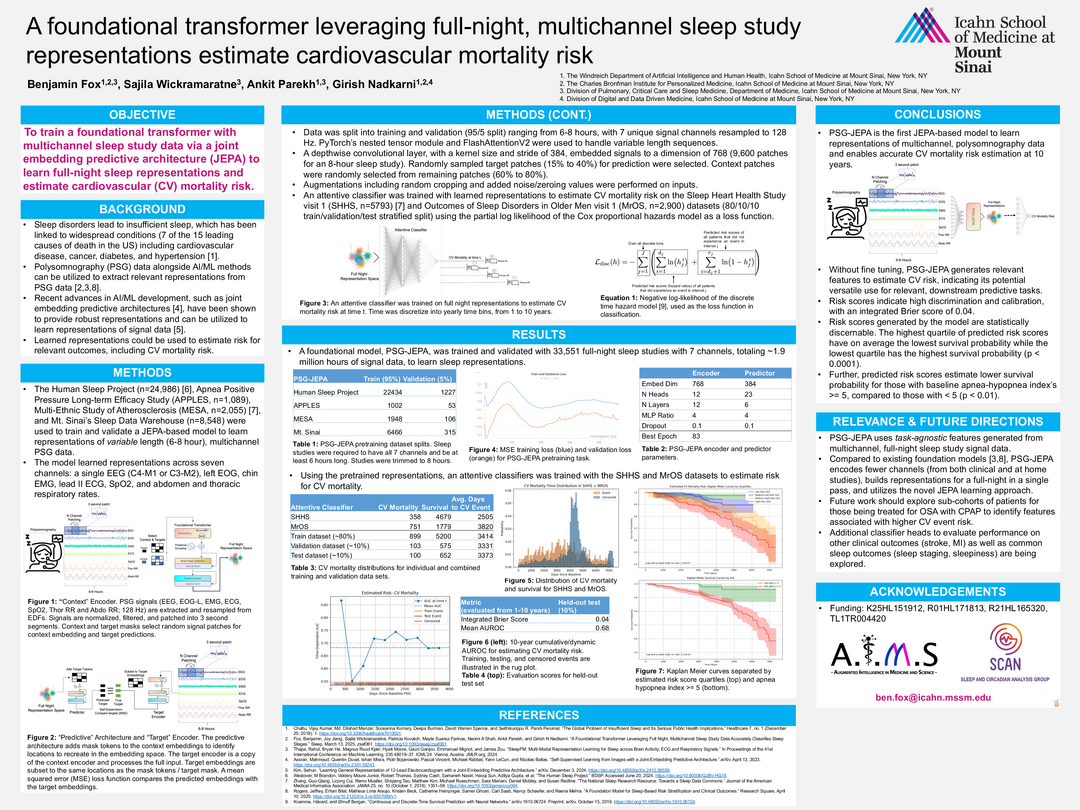
Title: A Foundational Transformer Leveraging Full-Night, Multichannel Sleep Study Representations to Estimate Cardiovascular Mortality Risk
Authors: Benjamin Fox, Sajila Wickramaratne, Ankit Parekh, Girish Nadkarni
Affiliations: Icahn School of Medicine at Mount Sinai
Objective:
This study introduces PSG-JEPA, a foundational transformer model trained on multichannel, full-night polysomnography (PSG) data using a Joint Embedding Predictive Architecture (JEPA). The model aims to learn robust, task-agnostic sleep representations to estimate 10-year cardiovascular (CV) mortality risk.
Background:
Sleep disorders are linked to several leading causes of death, including cardiovascular disease. PSG data, when combined with advanced AI/ML techniques, can yield meaningful representations for clinical risk prediction. JEPA, a recent self-supervised learning approach, offers a promising framework for learning from complex signal data.
Methods:
The model was trained on 33,551 PSG studies (~1.9 million hours) from four large datasets: the Human Sleep Project, APPLES, MESA, and Mt. Sinai’s Sleep Data Warehouse. Each study included seven signal channels (EEG, EOG, EMG, ECG, SpO₂, thoracic and abdominal respiratory rates), resampled to 128 Hz and segmented into 3-second patches. A depthwise convolutional encoder embedded these into a 768-dimensional space. JEPA was used to predict masked target patches from context patches using a mean squared error loss.
An attentive classifier was trained on the learned representations using the SHHS and MrOS datasets (n=8,693) to estimate CV mortality risk, with time discretized into yearly bins over a 10-year horizon. The Cox proportional hazards model was used as the loss function.
Results:
PSG-JEPA achieved a mean AUROC of 0.68 and an integrated Brier score of 0.04 on the held-out test set.
Risk scores showed strong calibration and discrimination. Kaplan-Meier curves demonstrated statistically significant survival differences across risk quartiles (p < 0.0001).
Individuals with an apnea-hypopnea index (AHI) ≥ 5 had significantly lower survival probabilities than those with AHI < 5 (p < 0.01).
The model generalized well without fine-tuning, indicating its potential for broader clinical applications.
Conclusions:
PSG-JEPA is the first JEPA-based model to learn full-night, multichannel PSG representations for CV mortality risk prediction. Its ability to generate meaningful, task-agnostic features without fine-tuning highlights its utility for downstream clinical tasks. Future directions include evaluating performance on other outcomes (e.g., stroke, MI, sleep staging) and exploring sub-cohorts such as CPAP-treated patients.
Funding: K25HL151912, R01HL171813, R21HL165320, TL1TR004420
Authors: Benjamin Fox, Sajila Wickramaratne, Ankit Parekh, Girish Nadkarni
Affiliations: Icahn School of Medicine at Mount Sinai
Objective:
This study introduces PSG-JEPA, a foundational transformer model trained on multichannel, full-night polysomnography (PSG) data using a Joint Embedding Predictive Architecture (JEPA). The model aims to learn robust, task-agnostic sleep representations to estimate 10-year cardiovascular (CV) mortality risk.
Background:
Sleep disorders are linked to several leading causes of death, including cardiovascular disease. PSG data, when combined with advanced AI/ML techniques, can yield meaningful representations for clinical risk prediction. JEPA, a recent self-supervised learning approach, offers a promising framework for learning from complex signal data.
Methods:
The model was trained on 33,551 PSG studies (~1.9 million hours) from four large datasets: the Human Sleep Project, APPLES, MESA, and Mt. Sinai’s Sleep Data Warehouse. Each study included seven signal channels (EEG, EOG, EMG, ECG, SpO₂, thoracic and abdominal respiratory rates), resampled to 128 Hz and segmented into 3-second patches. A depthwise convolutional encoder embedded these into a 768-dimensional space. JEPA was used to predict masked target patches from context patches using a mean squared error loss.
An attentive classifier was trained on the learned representations using the SHHS and MrOS datasets (n=8,693) to estimate CV mortality risk, with time discretized into yearly bins over a 10-year horizon. The Cox proportional hazards model was used as the loss function.
Results:
PSG-JEPA achieved a mean AUROC of 0.68 and an integrated Brier score of 0.04 on the held-out test set.
Risk scores showed strong calibration and discrimination. Kaplan-Meier curves demonstrated statistically significant survival differences across risk quartiles (p < 0.0001).
Individuals with an apnea-hypopnea index (AHI) ≥ 5 had significantly lower survival probabilities than those with AHI < 5 (p < 0.01).
The model generalized well without fine-tuning, indicating its potential for broader clinical applications.
Conclusions:
PSG-JEPA is the first JEPA-based model to learn full-night, multichannel PSG representations for CV mortality risk prediction. Its ability to generate meaningful, task-agnostic features without fine-tuning highlights its utility for downstream clinical tasks. Future directions include evaluating performance on other outcomes (e.g., stroke, MI, sleep staging) and exploring sub-cohorts such as CPAP-treated patients.
Funding: K25HL151912, R01HL171813, R21HL165320, TL1TR004420
Authors
First Author
Benjamin Fox, PhD
Contributing Authors
Sajila Wickramaratne, PhD
Ankit Parekh, PhD
Girish Nadkarni, MD, MPH
Poster